Brain Imaging Generation with Latent Diffusion Models
Notes
- The generated synthetic dataset is available at Academics Torrents
- A re-implementation is available at the following Monai link
Highlights
- Application of the Latent Diffusion Model framework for MR image synthesis of the human brain
- An encoder/decoder model dedicated to brain MRI reconstruction is proposed
-
Investigation of the application of conditioning to age, gender, ventricular volumes and brain volumes
- The model achieves new SOTA for brain MR image synthesis
- A synthetic dataset of 100,000 volumes, along with the conditioning information, is publicly available
Introduction
-
The objective of the paper is to generate a realistic large scale dataset with additional related “low dimensional” information such as age, sex or volumes.
-
31,740 T1w 3D MR images from the UK Biobank datas are used during training
-
One interest of such a dataset would be to provide enough data to learn to retrieve the age of a patient based on their brain MR image while guaranteeing privacy.
Methodology
Preprocessing steps
-
An existing network called UnitRes was used to perform a rigid body registration to a common MNI space
-
The final images are resampled to a uniform resolution of \(1 \, mm^3\)
-
The images are all cropped to a consistent volume size of \(160 \times 224 \times 160\) voxels
LDM architecture
- The method is directly inspired by the latent diffusion model whose architecture is summarized below:
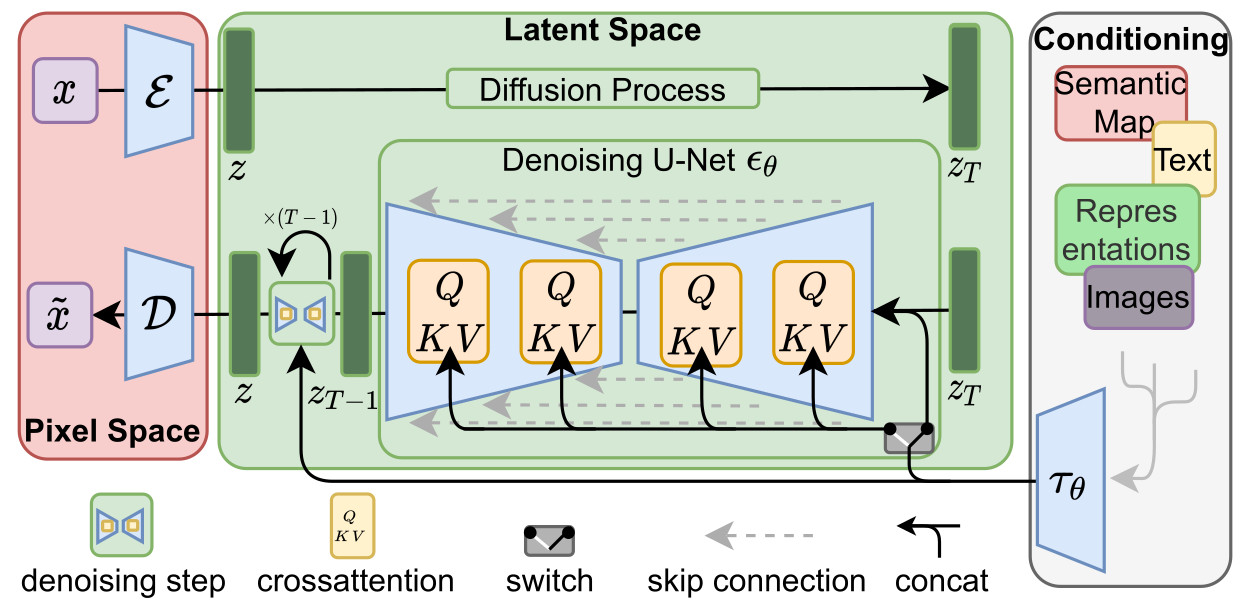
-
The autoencoder is first trained with a combination of L1 loss, perceptual loss, a patch-based adversarial objective and a KL regularization of the latent space
-
The encoder maps the brain image to a latent representation with a size of \(20 \times 28 \times 20\) voxels
-
The diffusion model is then trained using \(1000\) steps for the Markov chain process
-
The model is conditioned according to age, gender, ventricular volume and brain volume
-
The conditioning is performed by combining the concatenation of the conditioning with the input data and the use of cross-attention mechanisms
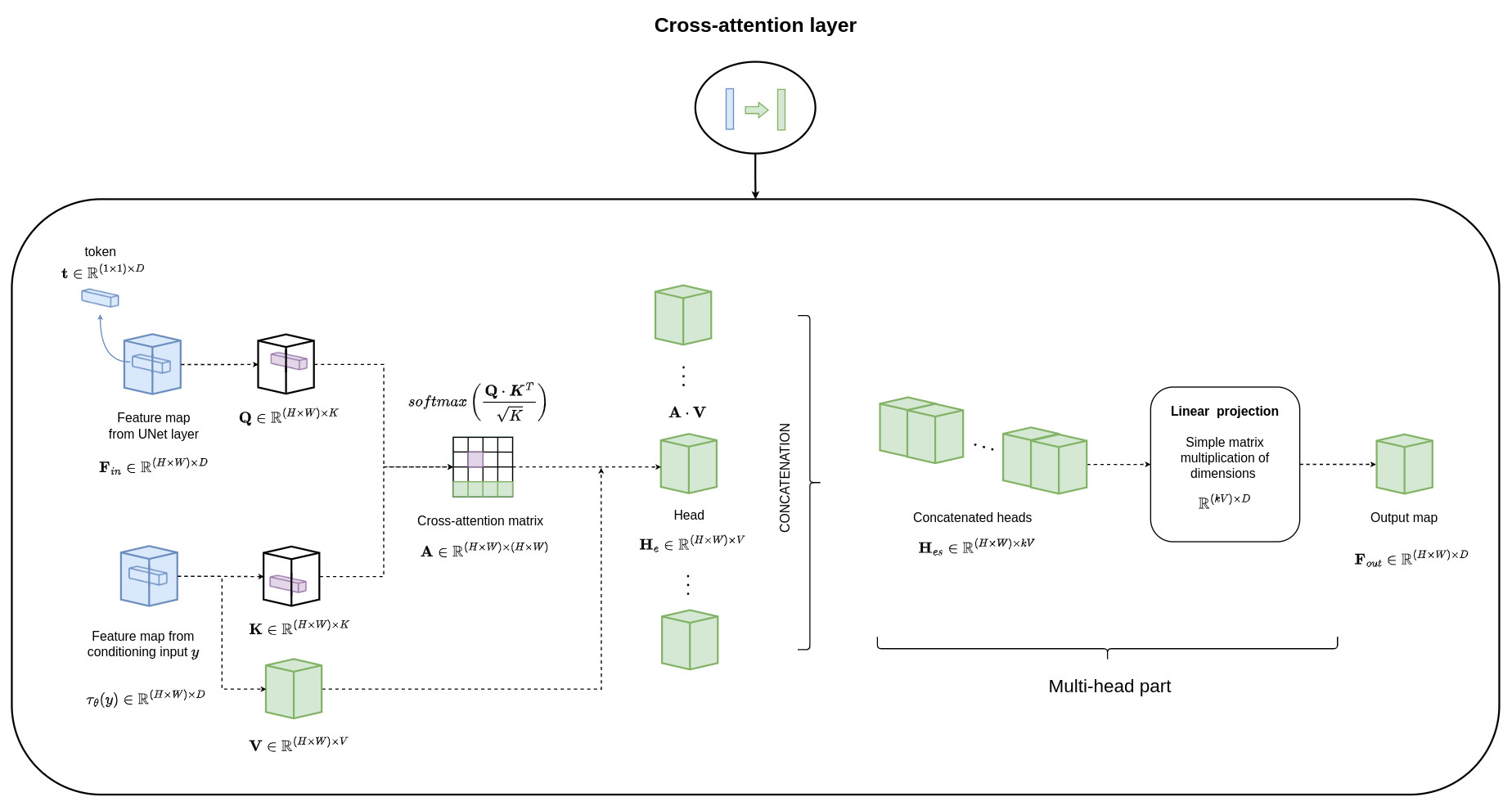
Results
- The autoencoder compressed each dimension of the input data by a factor of 8
- DDIM is used during inference to reduce from \(1000\) to \(50\) the number of time steps during sampling. This reduces the average sampling time from \(142 \pm 1.6\)s to \(7.6 \pm 0.2\)s
- The degree of realism of the synthetic data is measured using the Fréchet Inception Distance(FID), and the diversity of the data is measured with the Multi-Scale Structural Similarity metric (MS-SSIM) and the 4-G-R-SSIM
Quality of the synthetic data
- Measures were computed from 1000 sample pairs from the UK Biobank and the synthetic data
- The model achieves new SOTA for brain MR image synthesis
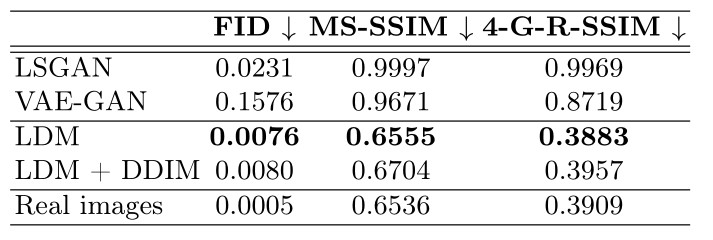
Figure 1. Quantitative evaluation of the synthetic images on the UK Biobank
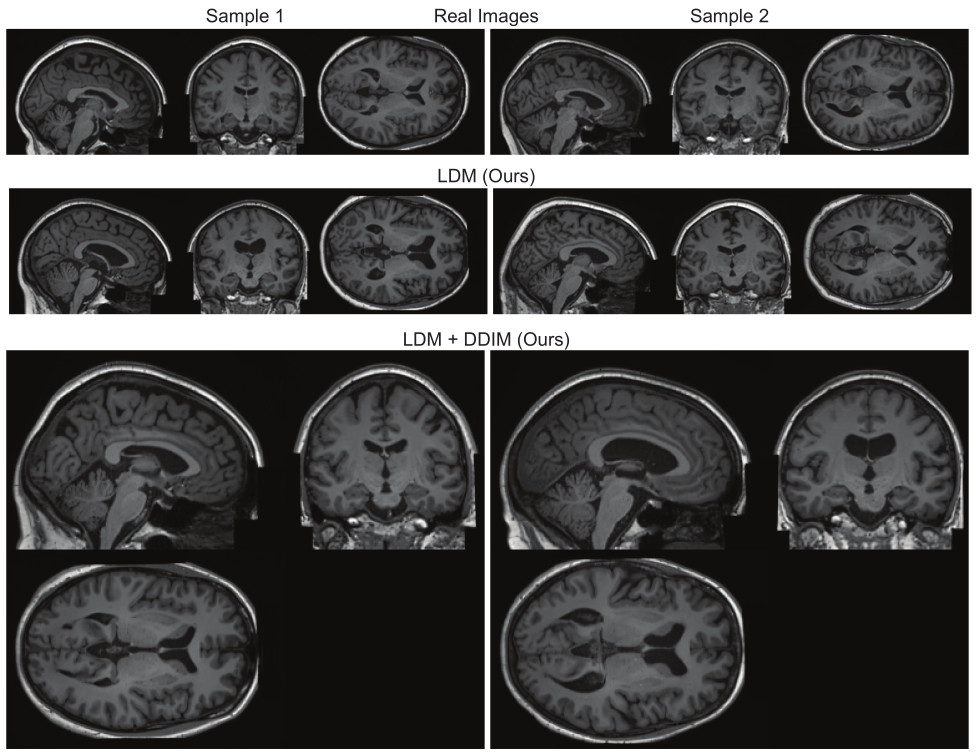
Figure 2. Real and synthetic samples of brain MRI
Conditioning on the ventricular volumes
-
To quantitatively evaluate the conditioning, SynthSeg was used to measure the volumes of the ventricles of 1000 synthetic brains
-
The Pearson correlation was computed between the obtained volumes and the inputted conditioning values
-
High correlation score of \(0.972\)
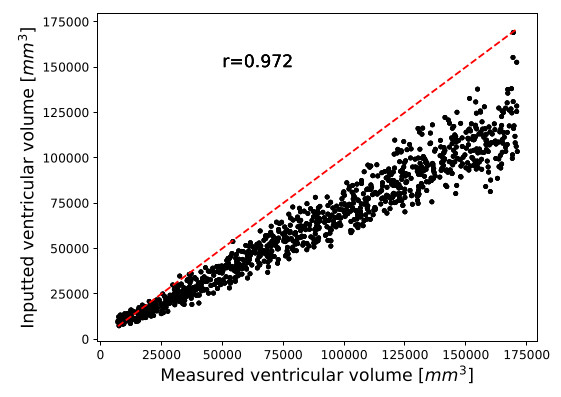
Figure 3. Correlation between inputted ventricular volumes and ventricular measured with SynthSeg
Conditioning on the age
-
A 3D CNN proposed in [1] was trained from the same UK Biobank dataset. The model takes as input a 3D brain image and predicts chronological age
-
The same model is then used on the synthetic dataset to verify how closely the predicted age matches the inputted age of the synthetic dataset
-
Good correlation score of \(0.692\)
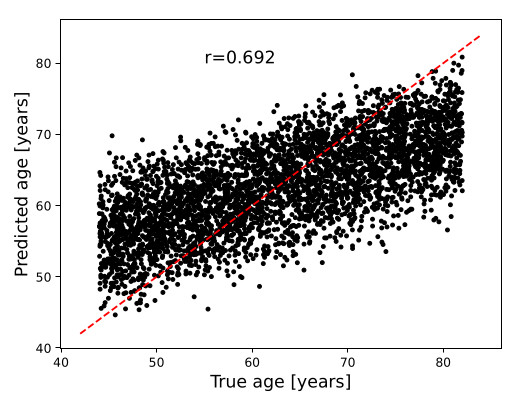
Figure 4. Correlation between inputted age and predicted brain age
Synthetic dataset
- A synthetic dataset of 100,000 human brain images was generated and made publicly available together with the conditioning information
Conclusions
- Latent diffusion model is cool ;)
- The key resides in the autoencoder performance !
- Is a database of 31,740 images really necessary ?
- We need to think carefully about the additive value of the conditioning information chosen to simulate a useful synthetic dataset !
References
[1] Cole, J.H., Poudel, R.P., Tsagkrasoulis, D., Caan, M.W., Steves, C., Spector, T.D., Montana, G., Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker, NeuroImage 163, 115–124 (2017)