I-MedSAM: Implicit Medical Image Segmentation with Segment Anything
Highlights
- The goal of the paper is to take advantage of both implicit representations and Segment Anything Model (SAM), to improve generalization and boundary delineation.
- The authors propose I-MedSAM, with a SAM-based encoder enriched by a frequency adapter, and a coarse-to-fine INR decoder with an uncertainty-guided sampling strategy.
Introduction
Standard segmentation methods (nnUNet, Transformers and more recently SAM) achieve effective results for this task but their discrete nature can lead to challenges. They are notably limited in their spatial flexibility.
Implicit Neural Representations (INRs) (or Neural Implicit Functions) are methods which learn a mapping from encoded image features and grid coordinates to the segmentation output. Those methods learn continuous representations and are therefore more flexible but they face challenges for domain transfer.
The goal of I-MedSAM is to leverage the benefits of SAM and INRs to improve generalization and boundary delineation.
Method
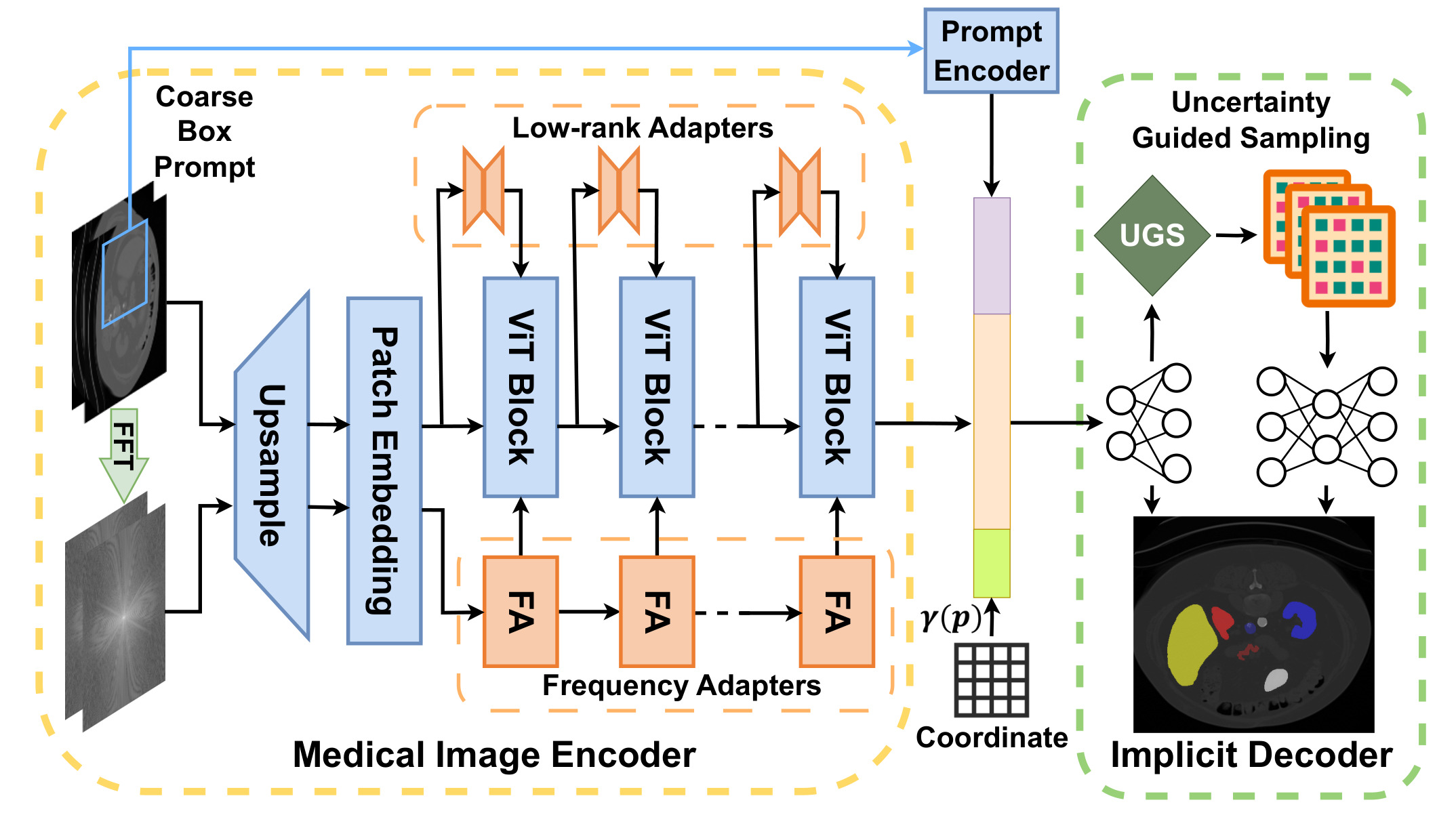
Figure 1: pipeline of I-MedSAM.
Medical Image Encoder
Main idea: Given a medical image and a prompt bounding box, multi-scale features are extracted from both spatial and frequency domains.
Similar to SAMed1, already presented in a previous post2, a Low-Rank Adapter (LoRA) is integrated into SAM. A novel Frequency Adapter (FA) is also integrated. This enables to do Parameter Efficient Fine Tuning (PEFT) and to update only a small number of parameters.
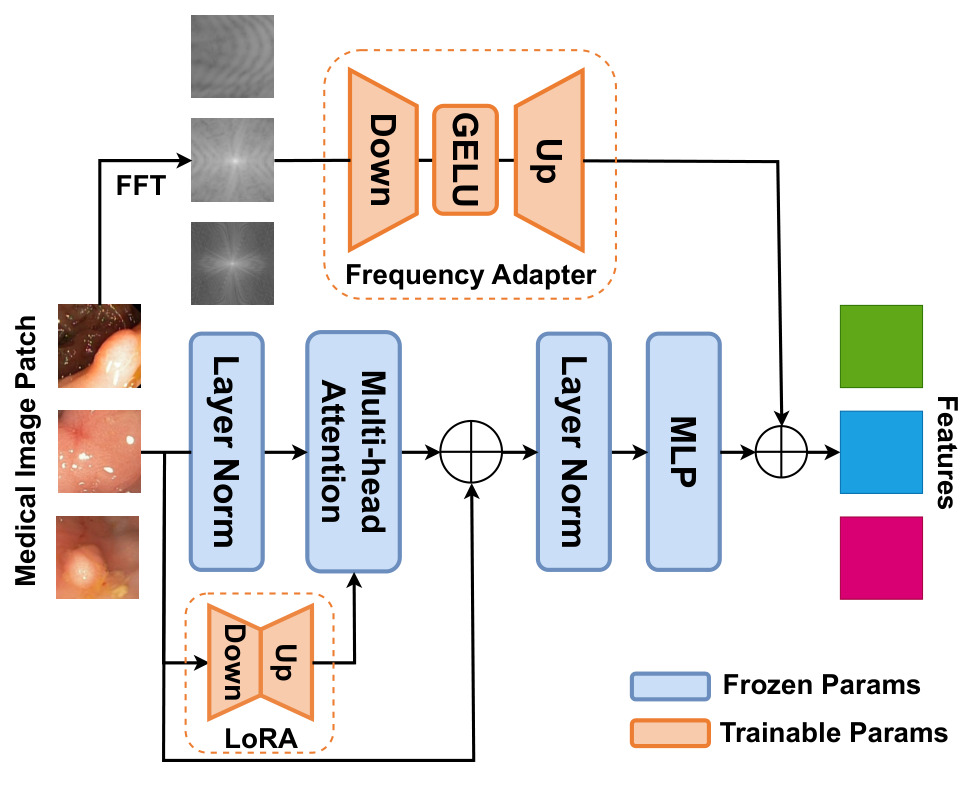
Figure 2: illustration of FA and LoRA in the image encoder.
Frequency Adapter (FA)
- present for each transformer block
- applied to the amplitude of the Fast Fourier Transform (FFT) of the image
- down-projection layer + GELU activation layer + up-projection layer
Low-Rank adapter (LoRA)
- present for each transformer block
- let \(W\) be the pre-trained weights, the update of \(W\) should be gradual and consistent: \(\hat{W} = W + \Delta W = W + BA\).
- LoRA is applied only on the Query and Value matrixes.
- rank = 4.
Implicit Segmentation Decoder
The input is a concatenation of:
- features from a coarse bounding box
- features from the image encoder interpolated to output resolution
- coordinates p mapped to higher dimensional space with \(\gamma(p)=(\sin(2^0\pi p),\cos(2^0\pi p),..., \sin(2^{L-1}\pi p),\cos(2^{L-1}\pi p) )\) where \(L = 10\).
Two INRs networks with a MLP architecture are optimized simultaneously.
- coarse/shallow INR : produce coarse segmentation \(\hat{o}^c\) and coarse features
- Uncertainty Guided Sampling (UGS): based on MC-dropout (Monte-Carlo dropout)
- dropout applied T times and gives T prediction results
- uncertainty for each pixel is the variance of the predictions
- points with the highest Top-K percentage are sampled = selected to be refined
- fine/deeper INR: using the coarse features as input, this network produces the fine segmentation probabilities \(\hat{o}^f\)
- Coarse and fine probabilities are combined to get the final segmentation map.
Training I-MedSAM
Loss:
- pixel-wise segmentation loss: \(L_{seg} = 0.5\times L_{ce}(o,\hat{o})+0.5\times L_{dice}(o,\hat{o})\)
- applied to supervise both coarse and fine segmentation maps
- during training, the weights for coarse supervision are slightly decreased until convergence
Datasets
Kvasir-Sessile
- 196 RGB images of small sessile polyps
- binary polyp segmentation
- Generalization capability evaluated on : CVC-ClinicDB (612 images from 31 colonoscopy sequences)
BCV
- 30 CT scans with annotations for 13 abdominal organs
- processed by slice
- Generalization capability evaluated on: AMOS (200 CT)
Results
Segmentation Comparison
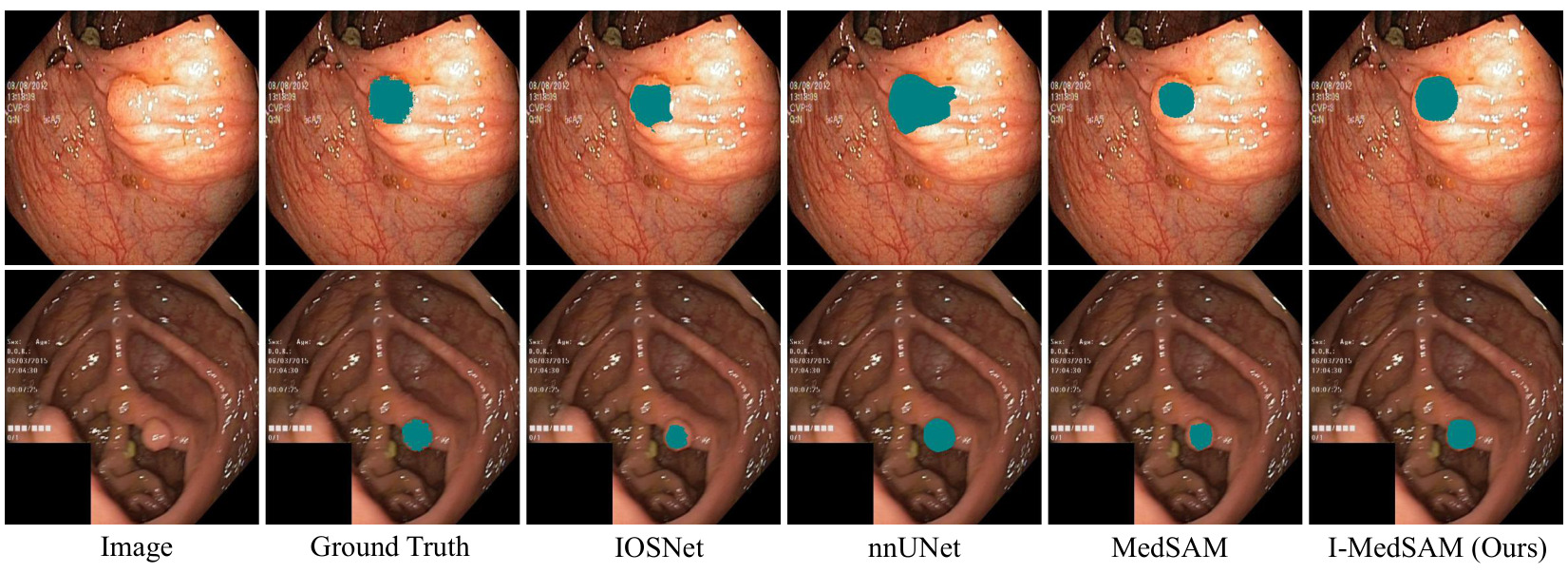 Figure 3: Qualitative results for Kvasir-Sessile dataset.
Figure 3: Qualitative results for Kvasir-Sessile dataset.
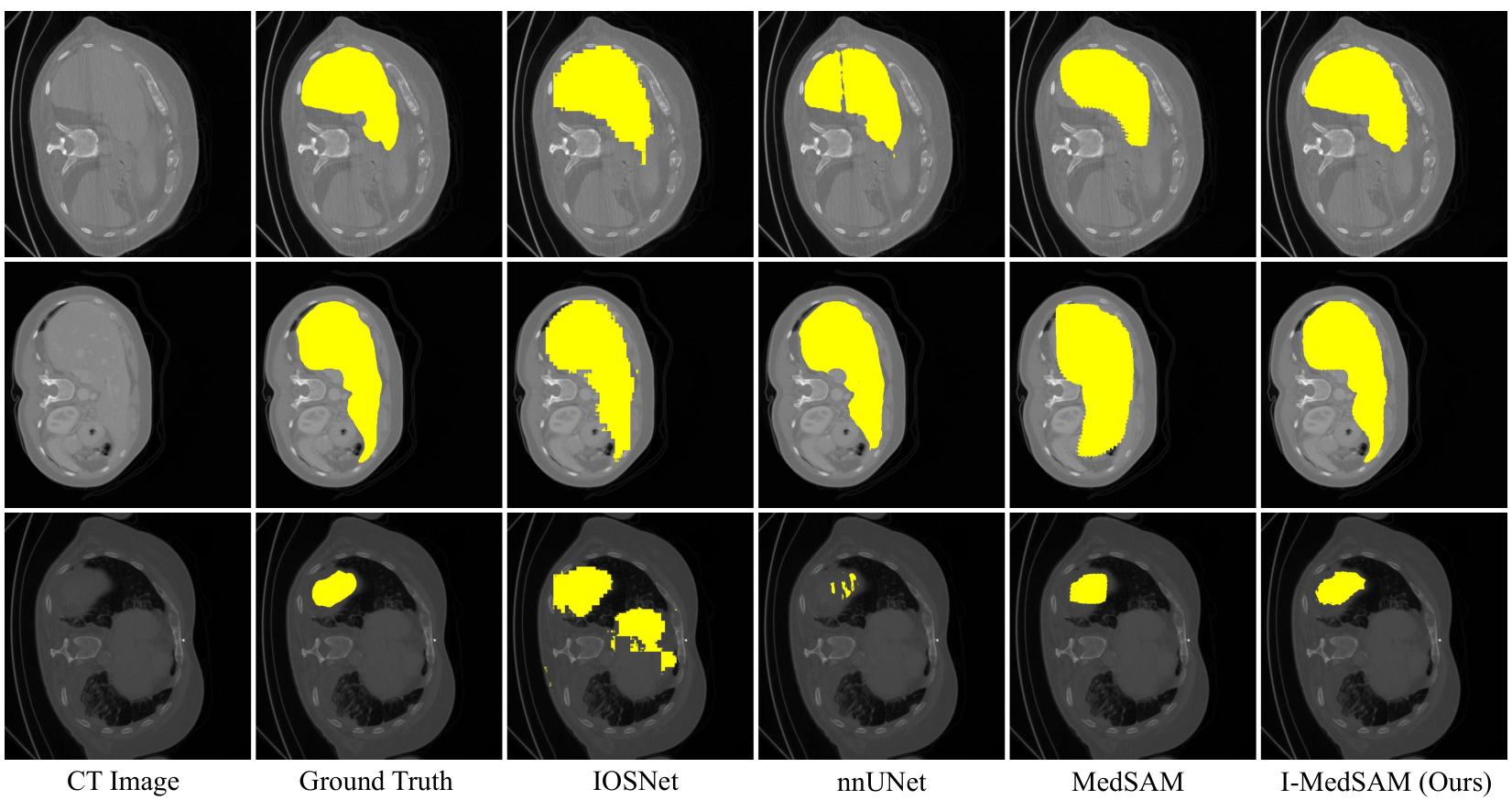 Figure 4: Qualitative results for BCV dataset.
Figure 4: Qualitative results for BCV dataset.
Visually, I-MedSAM gives better segmentation boundaries.
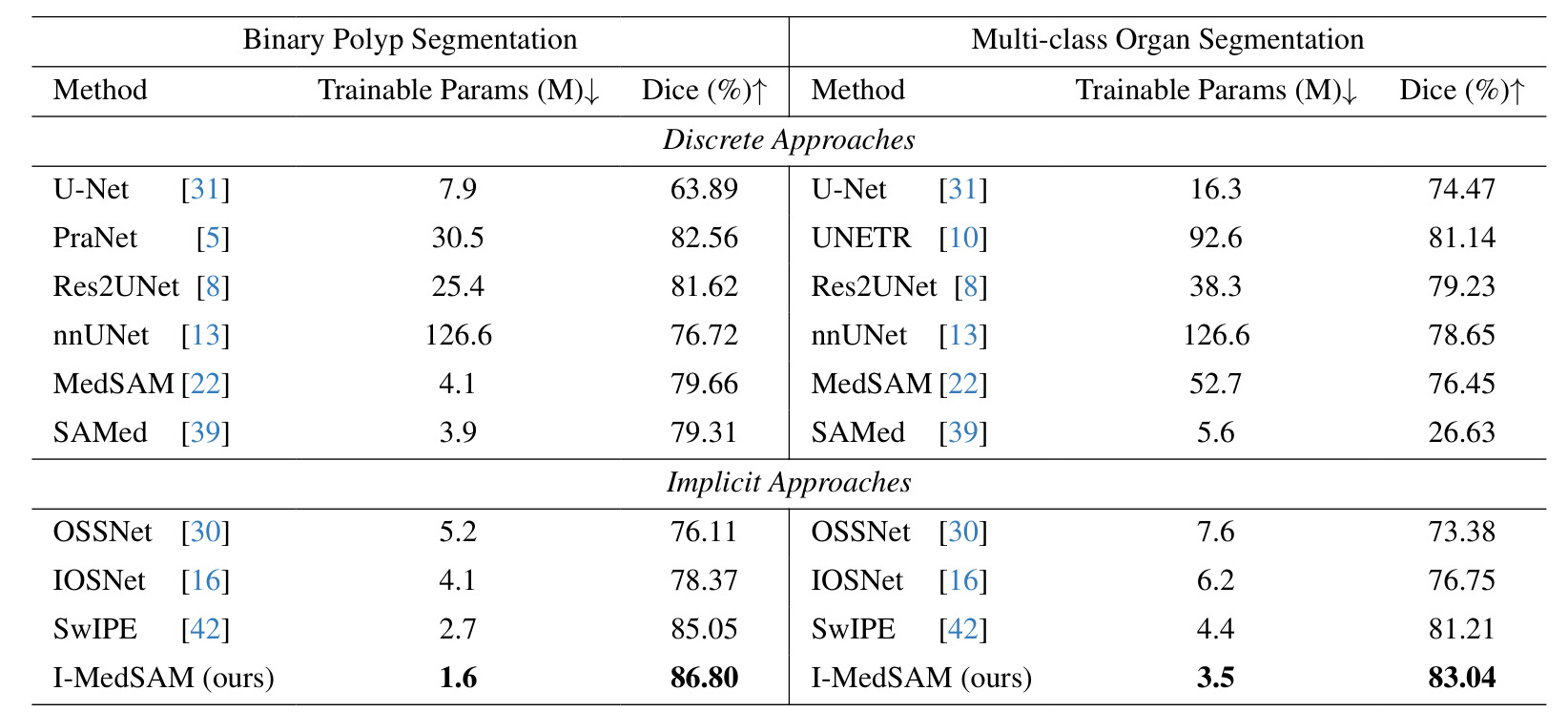 Table 1: Overall segmentation results.
Table 1: Overall segmentation results.
Results:
- Smaller KS dataset: notable improvements over implicit and discrete approaches
- BCV: I-MedSAM outperforms compared approaches but its improvements are less significant
Robustness under Data Shifts
Across resolutions
Trained and tested on different resolutions.
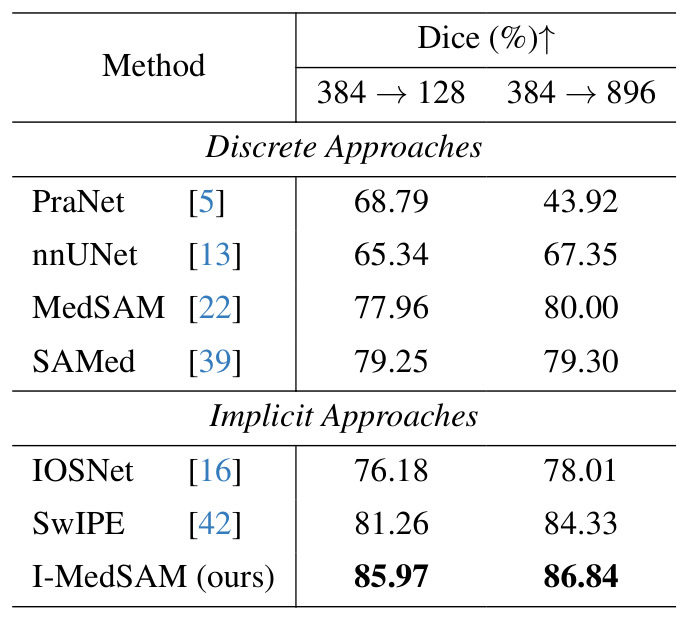 Table 2: Robustness across resolutions on Kvasir-Sessile dataset.
Table 2: Robustness across resolutions on Kvasir-Sessile dataset.
Results: implicit methods are more spatially flexible than discrete methods.
Across datasets
| Polyp segmentation | Organ segmentation |
|---|---|
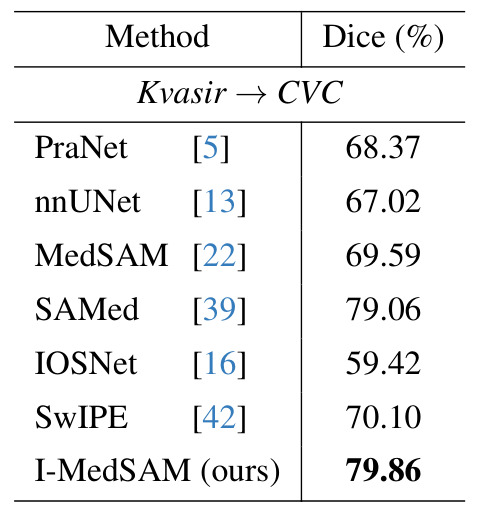 |
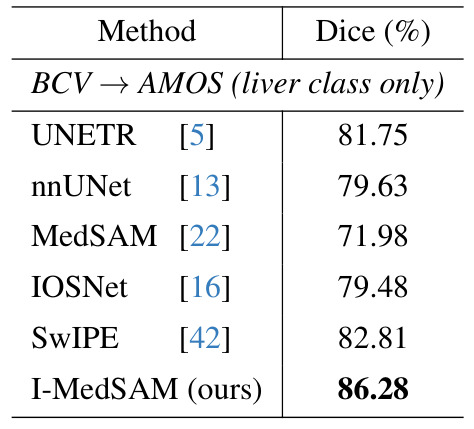 |
Results: I-MedSAM outperforms the best discrete methods.
Ablation study

Table 3: LoRA ranks.
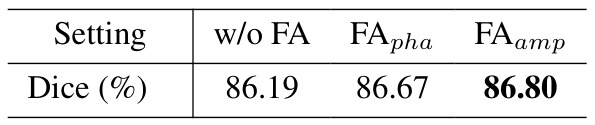
Table 4: Frequency Adapter: \(FA_{amp}\) uses amplitude and \(FA_{pha}\) uses phase
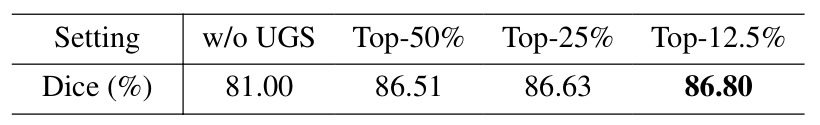 Table 5: Uncertainty Guided Sampling
Table 5: Uncertainty Guided Sampling
Using more points does not increase the dice results and requires more memory usage.
Conclusion
- With I-MedSAM, SAM with FA helps to learn better segmentation boundaries.
- Improvement of the method could be to develop an adapter to process different modalities.