UNeXt: MLP-based Rapid Medical Image Segmentation Network
Notes
- Link to the code here
Highlights
- The goal of the paper is to propose a segmentation network with a small number of parameters and computational complexity which could be implemented in point-of-care applications.
- The authors propose UNeXt, a UNet-based network which uses tokenized MLPs in the latent space to reduce the number of parameters.
Introduction
While medical image segmentation solutions like UNet and transformer-based networks have worked to improve segmentation performances, they haven’t focused on the computational cost of the algorithms.
This can be a problem for point-of-care imaging, which goal is to test and analyze images at the side of the patient. For example, skin conditions could be detected through a phone camera, and pleural irregularities and other conditions could be checked with point-of-care ultrasound (POCUS).
Those devices might not be able to deal with a high computation overhead, and therefore there is a need to develop performant and light algorithms that don’t require GPUs to run.
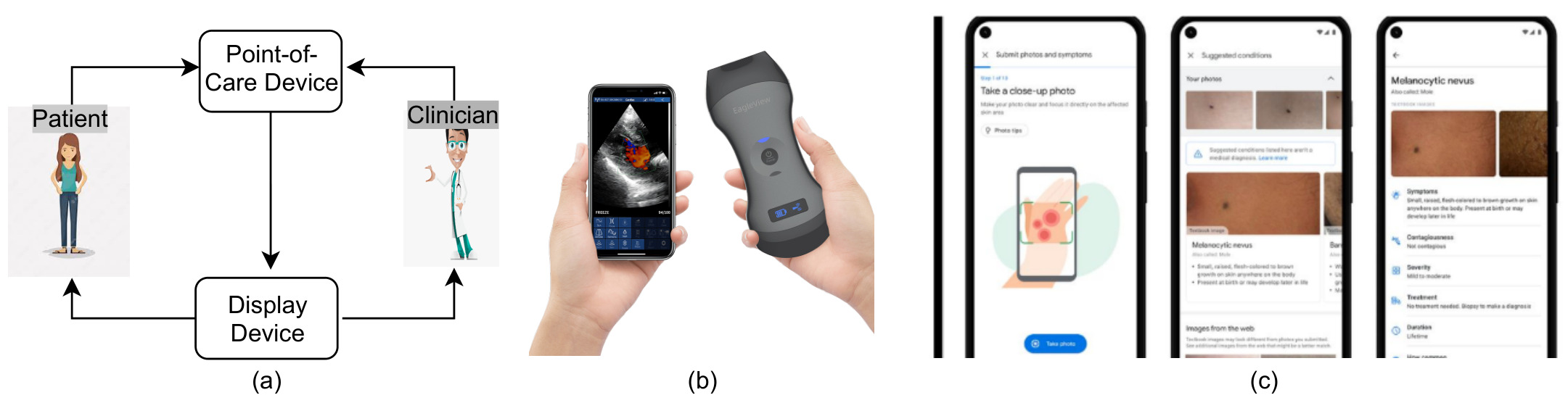
Figure 1: (a) Point-of-Care medical intervention workflow. (b) Recent medical imaging developments: POCUS device 1 and (c) Phone-based skin lesion detection and identification application 2.
Method
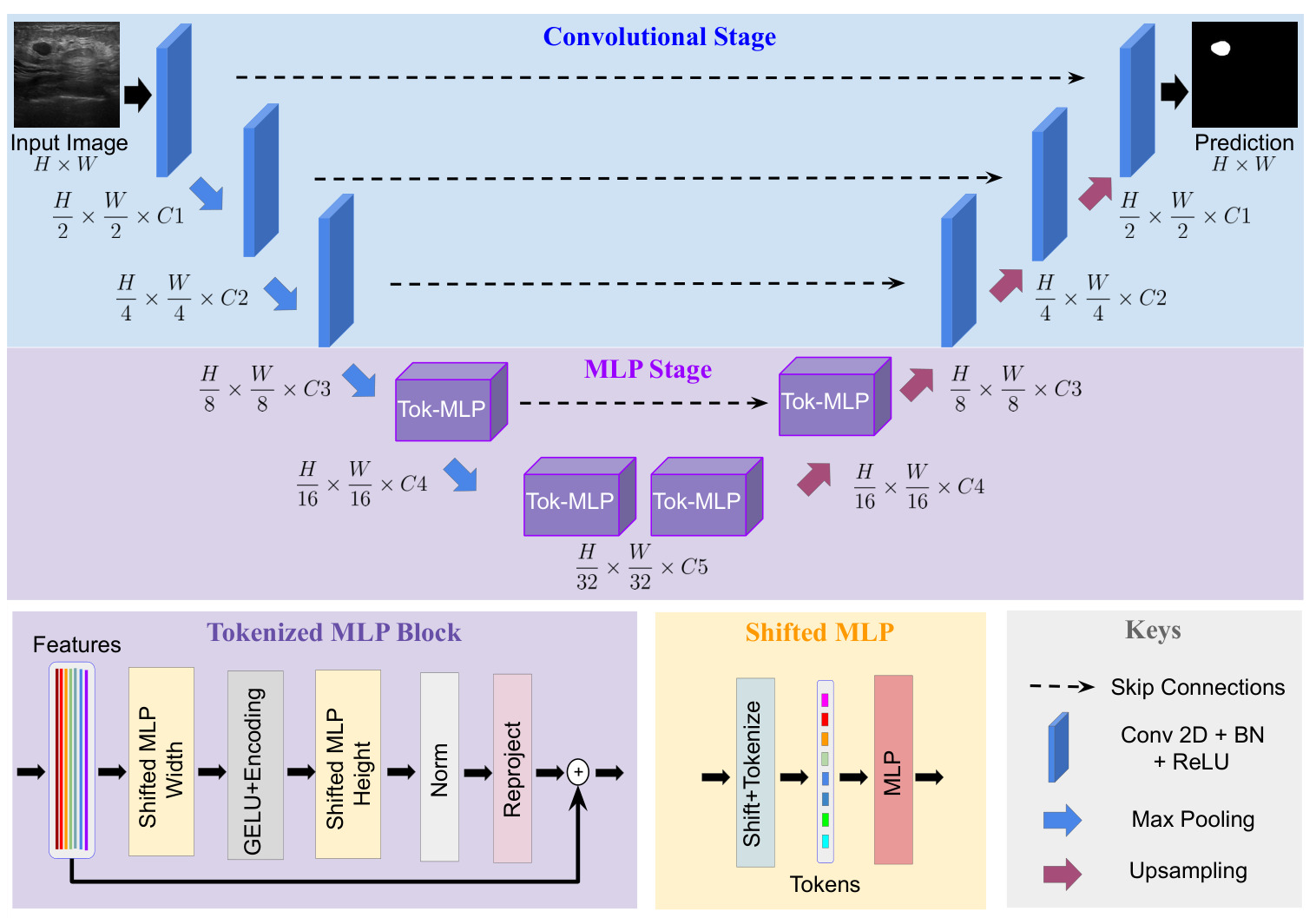
Figure 2: Architecture of UneXt.
Network Design
Architecture:
- UNet-like with encoder-decoder and skip-connections
- two stages: convolutional stage and tokenized MLP stage
Note: at each stage, the number of channel is reduced compared to UNet
Convolutional stage
- 3 convolutional blocks
- convolution layer, a batch normalization layer and ReLU activation
Tokenized MLP Stage
For each Tokenized MLP blocks:
1) A shift along the width axis is applied to the input feature map (see Figure 4).
2) The shifted feature maps are tokenized through a convolution layer.
3) An MLP processes the tokens
4) The output of the MLP is reshaped to 2D feature maps and processed by a depth wise (DW) convolutional layer. Here the idea is that a convolutional layer in an MLP block helps to do positional encoding (PE) i.e. helps to encode a positional information of the MLP features, and that a DW convolution requires less parameters than a regular convolution. This layer is followed by a GELU activation layer
5) The computed feature maps are shifted along the height, tokenized, and processed through a MLP.
6) The original tokens are added as residuals
7) The output is processed through a normalization layer and reshaped as 2D maps.
A depthwise convolution applies a single convolutional filter for each input channel. Each channel is kept separate.
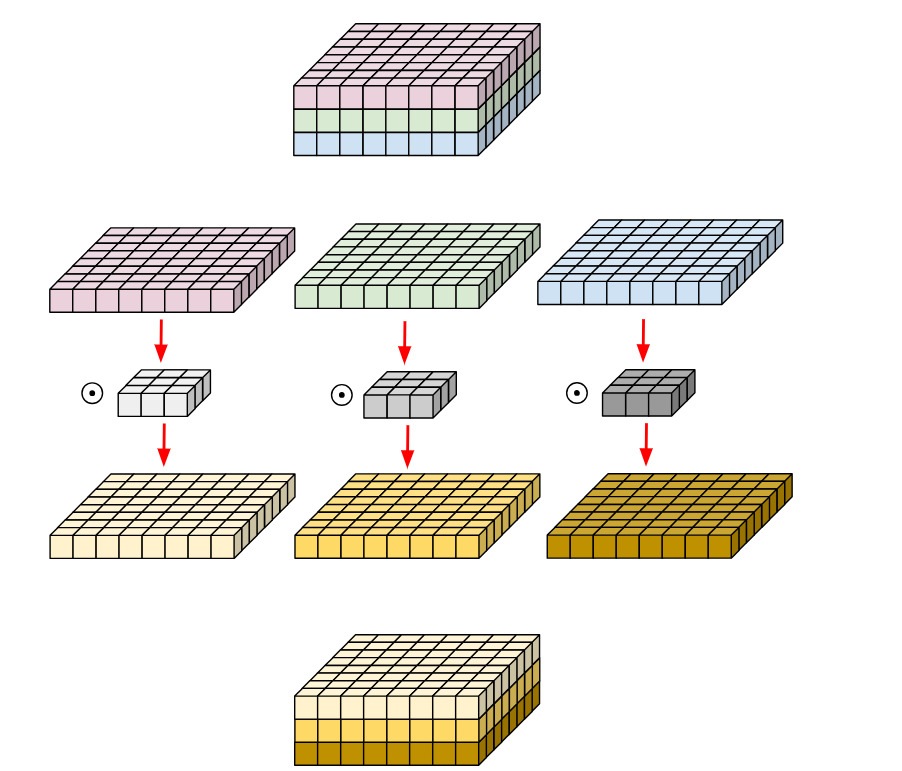
Figure 3: Depth-wise convolution, from 3
Shifted MLP
The authors state that shifting the window along the two axes introduces locality along them, and compare it with the window-based attention from the Swin transformers. The reader can also note that the shifting used is cyclic (torch.rolling used in the code).
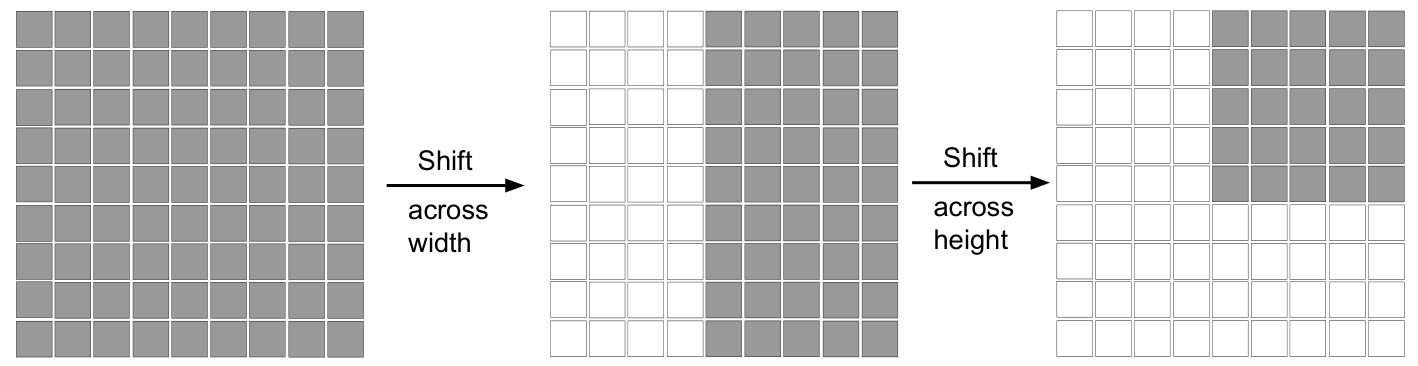
Figure 4: Shifting operation.
Implementation
- The loss used is a combination of binary cross-entropy loss and dice loss: \(L = 0.5\times BCE(\hat{y},y)+Dice(\hat{y},y)\)
- Number of tokens = 768
- Maximum shifting = 5 pixels
Other training details:
- Architecture implemented in PyTorch
- Adam Optimizer, learning rate of 0.0001
- 400 epochs
- Cosine annealing learning rate scheduler
Datasets
International Skin Imaging Collaboration (ISIC 2018)
- 2594 camera-acquired dermatologic images
- binary segmentation of skin lesions
Breast UltraSound Images (BUSI)
- 647 ultrasound images of normal, benign and malignant cases of breast cancer
- binary segmentation of benign and malignant images
Results
The authors repeat the trainings on three random splits and report the mean results here. The studied metrics are:
- F1 and IoU, to evaluate the segmentation performance
- the number of parameters
- the number of operations in GFLOPs
- the inference time on a CPU averaged for 10 images
Main Segmentation results

Figure 5: Row 1 - ISIC dataset, Row 2 - BUSI dataset. (a) Input. Predictions of (b) UNet (c) UNet++ (d) MedT (e) TransUNet (f) UNeXt and (g) Ground Truth.
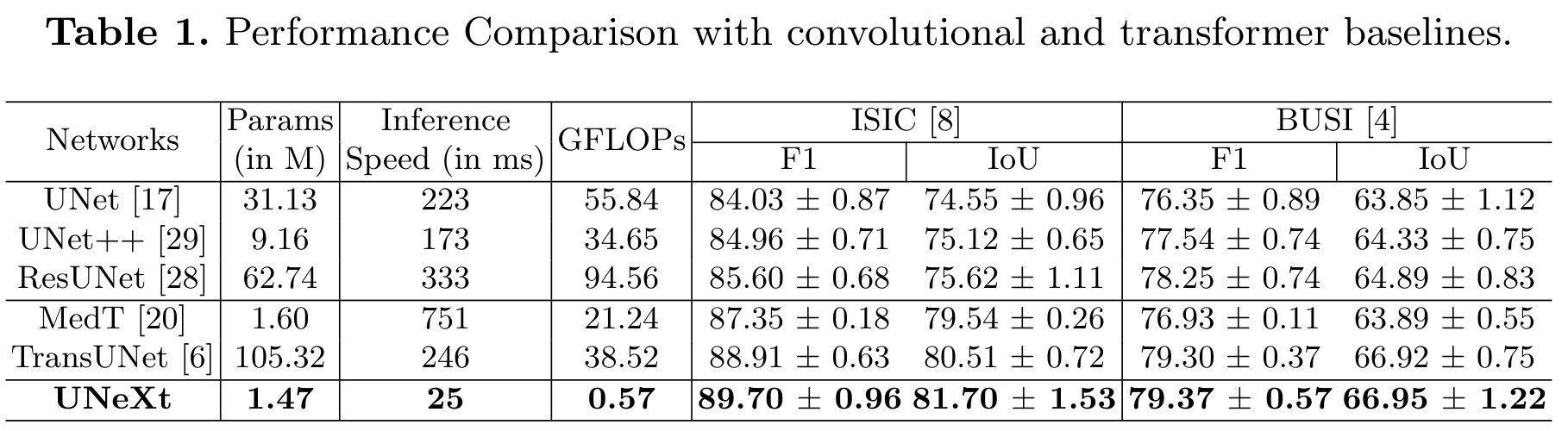
UNeXt has:
- better segmentation performance
- smallest number of parameters, smallest GFLOPs, fastest inference on the CPU
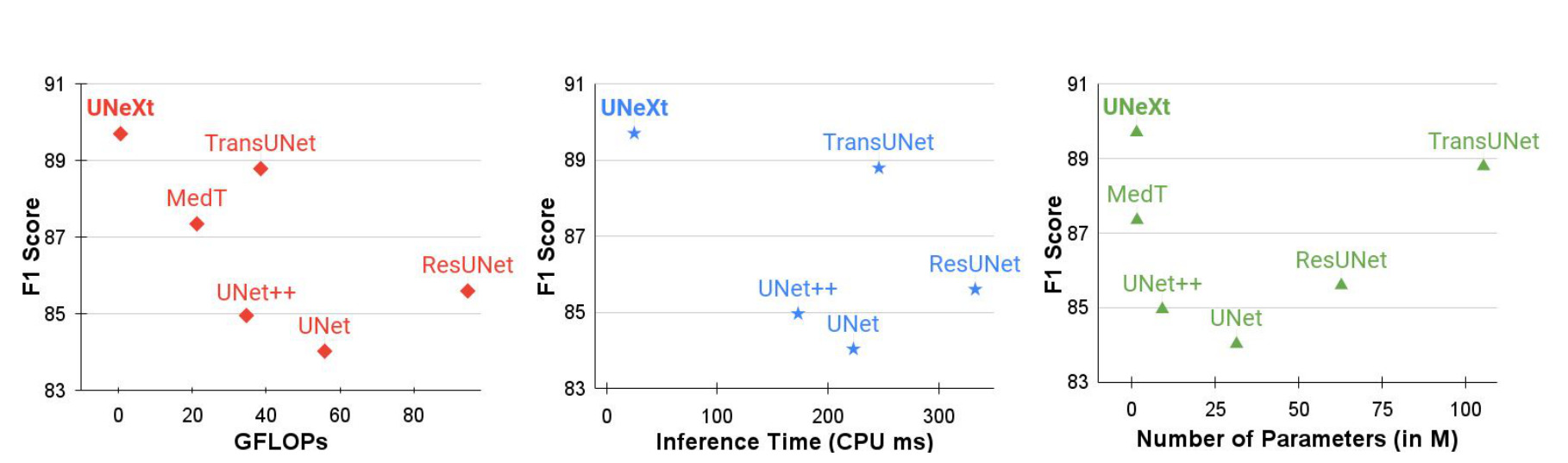
Figure 6: F1 score and GFLOPs, inference time or number of parameters for different networks.
Ablation study
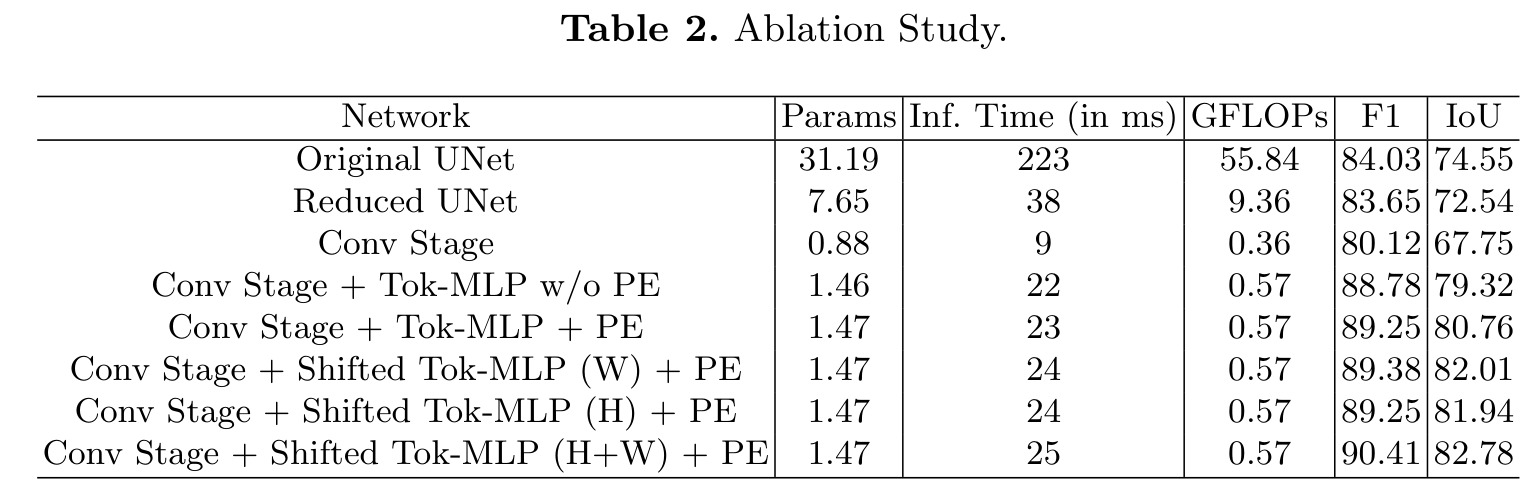
The goal here is to understand the individual contribution of each module:
- Original UNet
- Reduced UNet: reduce the number of filters
- Conv Stage: keep only 3 convolutional levels
- Conv Stage + Tok-MLP w/o PE : add the MLP blocks
- Conv Stage + Tok-MLP + PE : adds the DW convolutions
- Conv Stage + Shifted Tok-MLP (W) + PE : add the shifting operations along the width
- Conv Stage + Shifted Tok-MLP (W) + PE : add the shifting operations along the height
- Conv Stage + Shifted Tok-MLP (W) + PE : add the shifting operations along the width and height
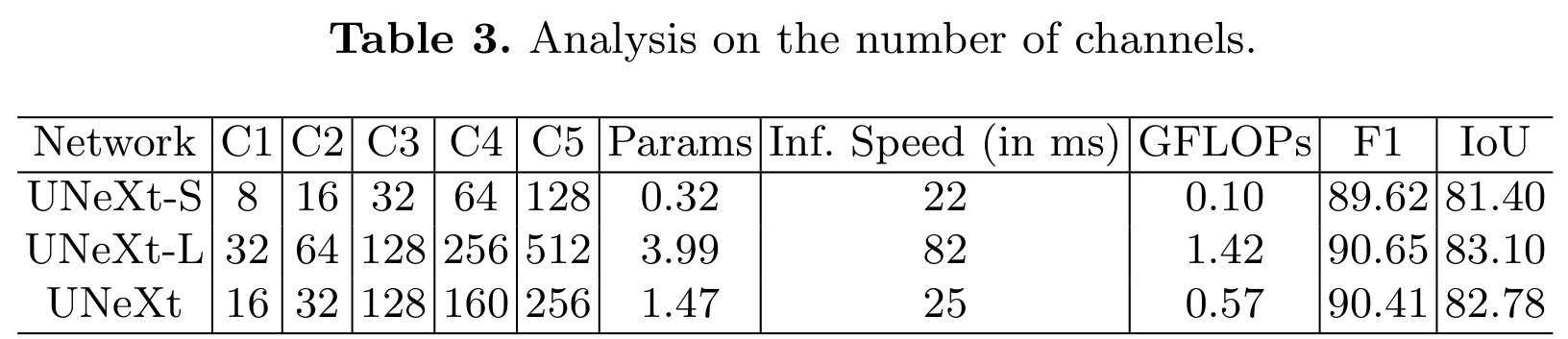
Analysis on number of channels
Study of the impact of the number of channels on the results of computation cost and segmentation performance, for a single fold of ISIC.
Difference from MLP-Mixer
Contrary to MLP-mixer, which is an all MLP architecture, UNeXt uses convolutional features and propose novel tokenized MLPs to model the represetation.
Conclusion
The proposed UNeXt reduces the number of parameters by 72x, decreases the computational complexity by 68x and increase the inference speed by 10x compared to the tranformer-based alternatives while keeping good segmentation performance.